import geopandas as gpd
import pandas as pd
import rasterio
from rasterio import windows
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import earthpy.plot as epI want to explore the vegetation of New South Wales in a couple of post using different packages and modules. I will start today with Python and my first question is how to work with a categorical map and its value attribute table.
Import the modules
First I use the import statements to load the modules I will use in this session. I will be using read functions from GeoPandas and Rasterio and a couple of helper funcitons from EarthPy as well as the usual suspects matplotlib, numpy and pandas.
The data
I will use the Vegetation Formations and Classes of NSW Version 3.03 (Keith and Simpson 2012; updated in 2017). The spatial data is available at the SEED portal, but I am too lazy to download and unzip the file myself, so I will just read the files on the fly.
Vegetation Formations and Classes of NSW (version 3.03 - 200m Raster) - David A. Keith and Christopher C. Simpson. VIS_ID 3848. Updated in 2017 as version 3.1. Available from SEED data portal
Value attribute table (VAT)
First, I want to read the value attribute table of the raster file (the file with the extension tif.vat.dbf). I will need to know the url for the zipfile, and then the relative path to the file that I want to read within the zipfile.
With these lines of code I generate the remote path I need for accessing the data:
url = "https://datasets.seed.nsw.gov.au/dataset/31986103-db62-4994-9702-054949281f56/resource/34a8cc72-5753-4993-8957-8d8af4fab008/download/ygeonetworkzipsvegetationnswmap3848.zip"
vat_file = "Vegetation_NSWmap3848/Data/GeoTIFF/NSWmap_v3_03_E_3848.tif.vat.dbf"
remote_path = 'zip+{}!/{}'.format(url, vat_file)I can read this file with geopandas:
vat = gpd.read_file(remote_path)And Now I can see the attribute table for the raster I want to use.
vat.head()| Value | Count | MapName | ClassNumbe | ClassName | FormationN | Formatio_1 | VIS_ID | Classifica | ThematicRe | SpatialRes | MapReliabi | Currency | Shape_Leng | Shape_Area | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2174.0 | Albury | 0 | Cleared | Cleared | 0.0 | 2907.0 | moderate | very coarse | 1:5000>scale>=1:25000, patches >=1 ha | Low | 2000-04 | 2.125684e+05 | 8.806694e+07 | None |
| 1 | 2 | 48.0 | Albury | 42 | Western Slopes Grassy Woodlands | Grassy woodlands | 4.0 | 2907.0 | moderate | very coarse | 1:5000>scale>=1:25000, patches >=1 ha | Low | 2000-04 | 5.519073e+04 | 2.160050e+06 | None |
| 2 | 3 | 1.0 | Albury | 52 | Inland Riverine Forests | Forested wetlands | 11.0 | 2907.0 | moderate | very coarse | 1:5000>scale>=1:25000, patches >=1 ha | Low | 2000-04 | 2.796665e+03 | 1.043730e+05 | None |
| 3 | 4 | 742.0 | Anabranch-Mildura | 52 | Inland Riverine Forests | Forested wetlands | 11.0 | 1873.0 | very low | coarse | scale>=1:100000, patches >=20 ha | Very low | 1964-1965 | 5.710281e+05 | 2.922206e+07 | None |
| 4 | 5 | 7817.0 | Anabranch-Mildura | 53 | Inland Floodplain Woodlands | Semi-arid woodlands (Grassy subformation) | 13.0 | 1873.0 | very low | coarse | scale>=1:100000, patches >=20 ha | Very low | 1964-1965 | 1.973462e+06 | 3.137011e+08 | None |
We will mostly use the class and formation names in columns ClassName and FormationN.
Raster data
Reading the GeoTiff file is very similar, I use the same url as before, but change the relative path to locate the file with .tif extension, then we open the remote path using rasterio.open:
gtiff_file = "Vegetation_NSWmap3848/Data/GeoTIFF/NSWmap_v3_03_E_3848.tif"
gtiff_path = 'zip+{}!/{}'.format(url, gtiff_file)
dataset = rasterio.open(gtiff_path)We can now explore this object to get its dimensions, bounds and coordinate reference system (CRS):
print(dataset.width, dataset.height)
{i: dtype for i, dtype in zip(dataset.indexes, dataset.dtypes)}
dataset.bounds
dataset.crs8661 5104CRS.from_epsg(3308)The CRS is important, for we will need this information to create a window to crop the raster data. This EPSG code is know as the GDA94 / NSW Lambert. I explore this projection in https://epsg.io/3308 and get the coordinates around Sturt National Park at the northwestern corner of New South Wales:
x0=8800000
y0=4900000
x1=x0+60000
y1=y0+40000Now I can use these bounds to crop the first (and only) band of the raster layer using the windows function in combination with a read function.
W = windows.from_bounds(
left=x0,
bottom=y0 ,
right=x1,
top=y1,
transform=dataset.transform)
band1 = dataset.read(1, window=W)We can now close the dataset, we already have the data we need in memory.
dataset.close()Summarising the data
Now we want to summarise the information over the selected spatial window. We can use the unique function from numpy to create a frequency table for the raster values present in this region of interest.
unique_values, counts = np.unique(band1, return_counts=True)
ss_vals = pd.DataFrame(index=unique_values, data={'NewCount':counts})And now we can join this with the VAT object created before. Make sure to use the right column for the index of both data frames to get the information aligned properly.
joined = vat.set_index('Value').join(ss_vals, how="right", validate='one_to_one')We can now create a summary table of the formations and classes present in our region of interest with the number of cells in each category.
joined.groupby(by=['FormationN','ClassName'])[['NewCount']].sum()| NewCount | ||
|---|---|---|
| FormationN | ClassName | |
| Arid shrublands (Acacia subformation) | Sand Plain Mulga Shrublands | 6858 |
| Stony Desert Mulga Shrublands | 870 | |
| Arid shrublands (Chenopod subformation) | Aeolian Chenopod Shrublands | 1432 |
| Gibber Chenopod Shrublands | 32782 | |
| Riverine Chenopod Shrublands | 170 | |
| Freshwater wetlands | Inland Floodplain Shrublands | 1523 |
| Semi-arid woodlands (Grassy subformation) | Wadi Woodlands | 15787 |
| Semi-arid woodlands (Shrubby subformation) | Desert Woodlands | 578 |
Plotting the data
We can plot this window of the map with a legend to look at the spatial configuration of these classes in the landscape.
First we choose the colors for our classes
clrs = plt.colormaps["tab20"](np.arange(len(unique_values)))
cmap=colors.ListedColormap(clrs)And we construct the legend combining the index number and the formation names:
class_names_list = list()
for a,b in zip(joined.index,joined.FormationN):
class_names_list.append("{} {}".format(a,b))And now we combine a couple of functions from matplotlib and EarthPy to create a plot with a legend:
f, ax = plt.subplots(figsize=(5,5))
im = ax.imshow(band1, cmap = cmap, norm=None)
ax.set(title="Formations")
ep.draw_legend(im, titles = class_names_list)
ax.set_axis_off()
plt.show()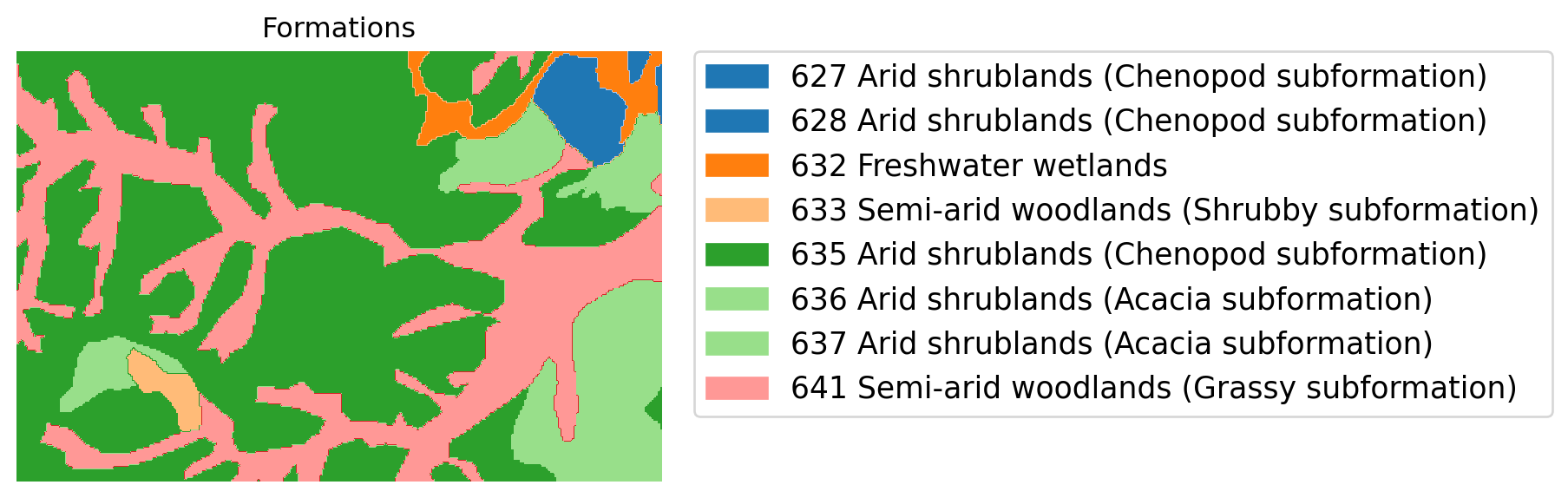
Conclusion
Here we use python, rasterio and geopandas to explore a categorical raster layer using its value attribute table. Thanks to NSW SEED portal for providing the data!
Here the basic recipe:
- Find the dataset url,
- Load the VAT file
- Load the raster layer and crop it to a region of interest
- Read the data and summarise it with numpy and pandas functions
- Plot the data with matplotlib
- Done!
We can now build on this in future posts. Cheers!